K-Means
K-Means#
K-Means 클러스터링은 초기 값으로 주어진 Centroid의 위치 및 갯수를 통해 가까운 거리의 점들을 클러스터 하고 Centroid 를 다시 계산하는 방식을 거져 Centroid 의 위치가 최적화 될때까지 클러스링을 진행합니다.
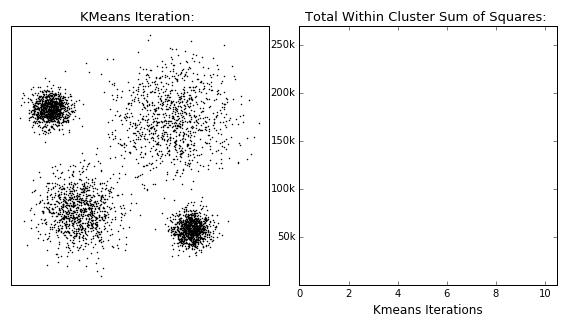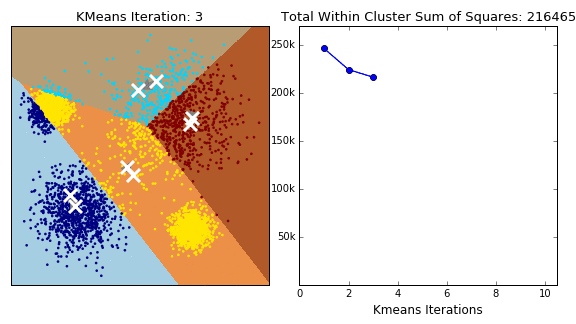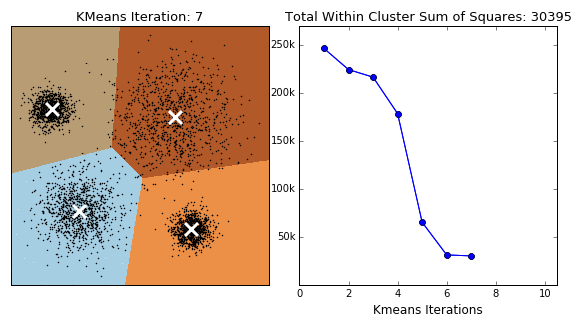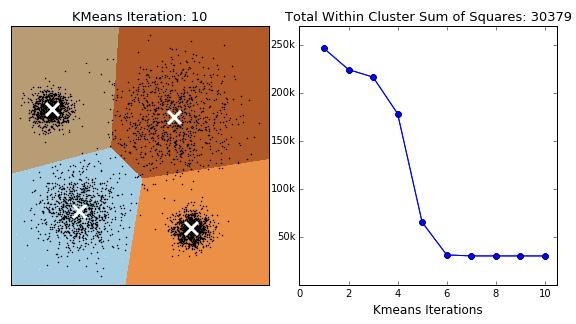
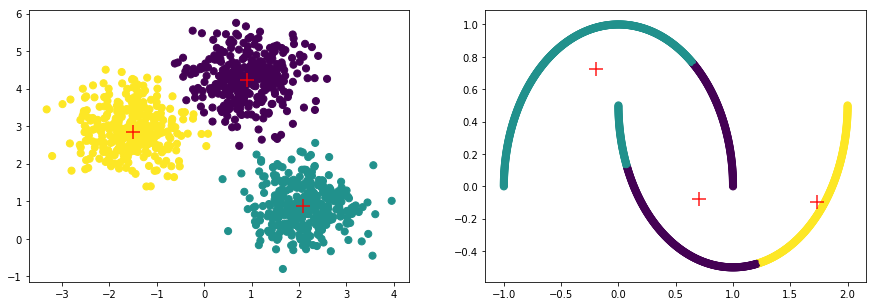
K-Means 클러스터링 방식은 클러스터의 군집, 밀도가 고르고 Circle 형태의 클러스터에서는 잘 작동하는 반면, 아래와 같은 Moon 형식의 데이터에서는 만족할 만한 성과를 내지 못합니다.
%matplotlib inline
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore")
import numpy as np
from sklearn import datasets, model_selection, cluster, metrics
# 데이터
np.random.seed(0)
n_samples = 1000
plt.rcParams['figure.figsize'] = (15,5)
np_datasets = []
np_data_xs, np_data_ys = datasets.samples_generator.make_blobs(
n_samples=n_samples,
centers=3,
cluster_std=0.60,
random_state=0)
print("data shape: np_data_xs={}, np_data_ys={}".format(np_data_xs.shape, np_data_ys.shape))
np_datasets.append((np_data_xs, np_data_ys))
plt.subplot(1, 2, 1)
plt.scatter(np_data_xs[:, 0], np_data_xs[:, 1], c=np_data_ys)
np_data_xs, np_data_ys = datasets.make_moons(
n_samples=n_samples,
random_state=0)
print("data shape: np_data_xs={}, np_data_ys={}".format(np_data_xs.shape, np_data_ys.shape))
np_datasets.append((np_data_xs, np_data_ys))
plt.subplot(1, 2, 2)
plt.scatter(np_data_xs[:, 0], np_data_xs[:, 1], c=np_data_ys)
plt.show()
# 모델
# MiniBatchKMeans: KMeans 보다 수행시간을 최적화한 버전
# init='k-means++': centroid 를 서로 떨어진 상태로 초기화하여 결과 향상에 도움을 줍니다.
models = [
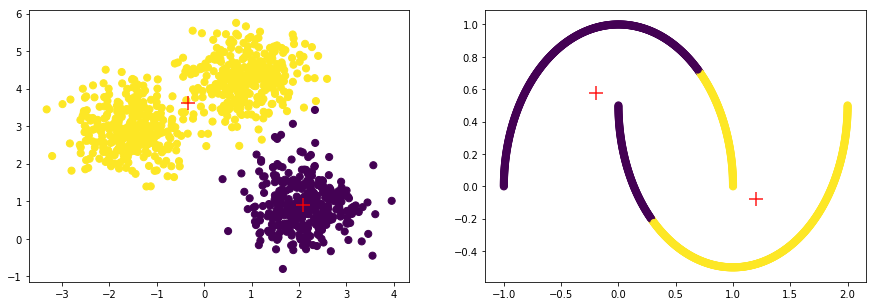
cluster.KMeans(init='k-means++', n_clusters=2),
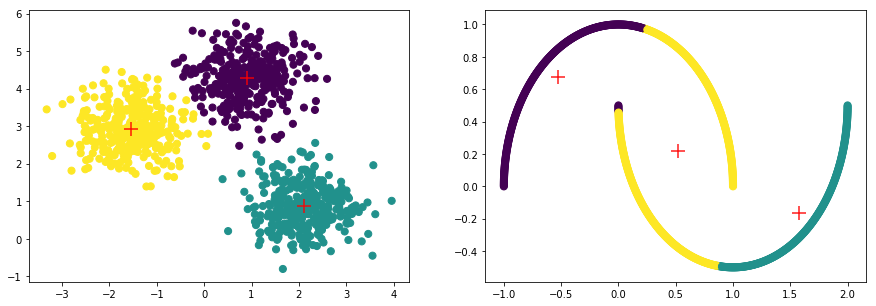
cluster.KMeans(init='k-means++', n_clusters=3),
cluster.MiniBatchKMeans(init='k-means++', batch_size=6, n_clusters=2),
cluster.MiniBatchKMeans(init='k-means++', batch_size=6, n_clusters=3),
]
for model in models:
for i, np_dataset in enumerate(np_datasets):
np_data_xs, np_data_ys = np_dataset[0], np_dataset[1]
# 학습
print("\nmodel={}".format(model))
model.fit(np_data_xs)
# 평가
labels = model.labels_
centers = model.cluster_centers_
plt.subplot(1, 2, i+1)
plt.scatter(np_data_xs[:, 0], np_data_xs[:, 1], c=labels, s=50, cmap='viridis')
plt.scatter(centers[:, 0], centers[:, 1], c='r', marker = '+', s=200, alpha=0.9);
score = metrics.silhouette_score(np_data_xs, labels, metric='euclidean')
print("score={:.5f}".format(score))
plt.show()
data shape: np_data_xs=(1000, 2), np_data_ys=(1000,)
data shape: np_data_xs=(1000, 2), np_data_ys=(1000,)
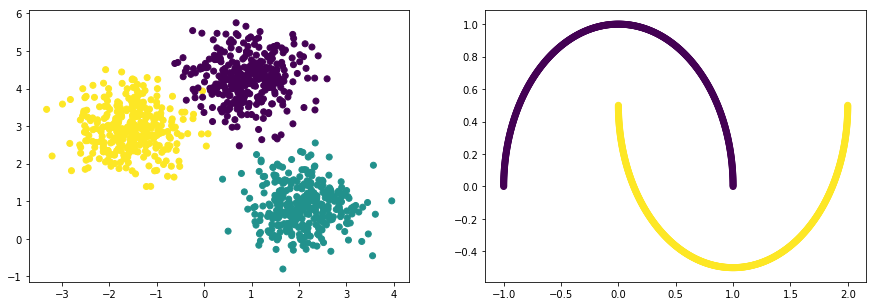
model=KMeans(algorithm='auto', copy_x=True, init='k-means++', max_iter=300,
n_clusters=2, n_init=10, n_jobs=None, precompute_distances='auto',
random_state=None, tol=0.0001, verbose=0)
score=0.57071
model=KMeans(algorithm='auto', copy_x=True, init='k-means++', max_iter=300,
n_clusters=2, n_init=10, n_jobs=None, precompute_distances='auto',
random_state=None, tol=0.0001, verbose=0)
score=0.49305
model=KMeans(algorithm='auto', copy_x=True, init='k-means++', max_iter=300,
n_clusters=3, n_init=10, n_jobs=None, precompute_distances='auto',
random_state=None, tol=0.0001, verbose=0)
score=0.65326
model=KMeans(algorithm='auto', copy_x=True, init='k-means++', max_iter=300,
n_clusters=3, n_init=10, n_jobs=None, precompute_distances='auto',
random_state=None, tol=0.0001, verbose=0)
score=0.43287
model=MiniBatchKMeans(batch_size=6, compute_labels=True, init='k-means++',
init_size=None, max_iter=100, max_no_improvement=10, n_clusters=2,
n_init=3, random_state=None, reassignment_ratio=0.01, tol=0.0,
verbose=0)
score=0.56947
model=MiniBatchKMeans(batch_size=6, compute_labels=True, init='k-means++',
init_size=None, max_iter=100, max_no_improvement=10, n_clusters=2,
n_init=3, random_state=None, reassignment_ratio=0.01, tol=0.0,
verbose=0)
score=0.49231
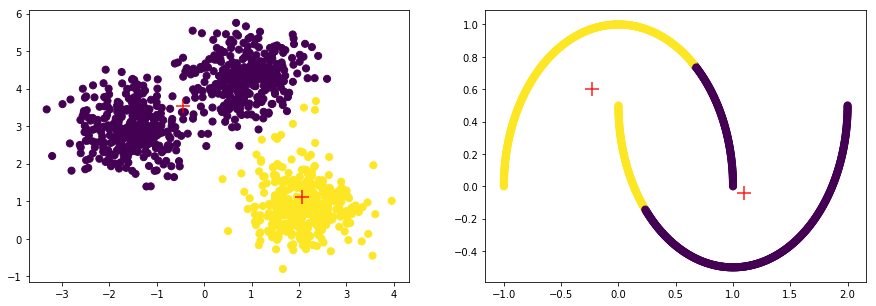
model=MiniBatchKMeans(batch_size=6, compute_labels=True, init='k-means++',
init_size=None, max_iter=100, max_no_improvement=10, n_clusters=3,
n_init=3, random_state=None, reassignment_ratio=0.01, tol=0.0,
verbose=0)
score=0.65324
model=MiniBatchKMeans(batch_size=6, compute_labels=True, init='k-means++',
init_size=None, max_iter=100, max_no_improvement=10, n_clusters=3,
n_init=3, random_state=None, reassignment_ratio=0.01, tol=0.0,
verbose=0)
score=0.43628