SVM Classification
SVM Classification#
SVM(Support Vector Machine) Classification
Support Vector Machine은 머신러닝 분야에서 우수한 알고리즘 중에 하나로 데이터 형태에 맞는 Kernel 함수 및 Regularization를 선택하여 적용함으로써, 선형/비선형 데이터셋 및 분류/회귀 문제 모두에 사용할수 있습니다.
SVM 은 Feature의 Scale 에 민감하므로 학습 전에 전처리를 통해 Feature 의 Sacale 을 조정해 줘야 합니다.
# 경고 메시지 출력 끄기
import warnings
warnings.filterwarnings(action='ignore')
%matplotlib inline
import matplotlib.pyplot as plt
import IPython
import platform, sys
rseed = 22
import random
random.seed(rseed)
import numpy as np
np.random.seed(rseed)
np.set_printoptions(precision=3)
np.set_printoptions(formatter={'float_kind': "{:.3f}".format})
import pandas as pd
pd.set_option('display.max_rows', None)
pd.set_option('display.max_columns', None)
pd.set_option('display.max_colwidth', None)
pd.options.display.float_format = '{:,.5f}'.format
import sklearn
print(f"python ver={sys.version}")
print(f"python platform={platform.architecture()}")
print(f"pandas ver={pd.__version__}")
print(f"numpy ver={np.__version__}")
print(f"sklearn ver={sklearn.__version__}")
python ver=3.8.9 (default, Jun 12 2021, 23:47:44)
[Clang 12.0.5 (clang-1205.0.22.9)]
python platform=('64bit', '')
pandas ver=1.2.4
numpy ver=1.19.5
sklearn ver=0.24.2
#참조: https://scikit-learn.org/stable/auto_examples/svm/plot_iris_svc.html
print(__doc__)
import numpy as np
import matplotlib.pyplot as plt
from sklearn import svm, datasets
def make_meshgrid(x, y, h=.02):
"""Create a mesh of points to plot in
Parameters
----------
x: data to base x-axis meshgrid on
y: data to base y-axis meshgrid on
h: stepsize for meshgrid, optional
Returns
-------
xx, yy : ndarray
"""
x_min, x_max = x.min() - 1, x.max() + 1
y_min, y_max = y.min() - 1, y.max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, h),
np.arange(y_min, y_max, h))
return xx, yy
def plot_contours(ax, clf, xx, yy, **params):
"""Plot the decision boundaries for a classifier.
Parameters
----------
ax: matplotlib axes object
clf: a classifier
xx: meshgrid ndarray
yy: meshgrid ndarray
params: dictionary of params to pass to contourf, optional
"""
Z = clf.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
out = ax.contourf(xx, yy, Z, **params)
return out
# import some data to play with
iris = datasets.load_iris()
# Take the first two features. We could avoid this by using a two-dim dataset
X = iris.data[:, :2]
y = iris.target
# we create an instance of SVM and fit out data. We do not scale our
# data since we want to plot the support vectors
C = 1.0 # SVM regularization parameter
models = (svm.SVC(kernel='linear', C=C),
svm.LinearSVC(C=C, max_iter=10000),
svm.SVC(kernel='rbf', gamma=0.7, C=C),
svm.SVC(kernel='poly', degree=3, gamma='auto', C=C))
models = (clf.fit(X, y) for clf in models)
# title for the plots
titles = ('SVC with linear kernel',
'LinearSVC (linear kernel)',
'SVC with RBF kernel',
'SVC with polynomial (degree 3) kernel')
# Set-up 2x2 grid for plotting.
fig, sub = plt.subplots(2, 2)
plt.subplots_adjust(wspace=0.4, hspace=0.4)
X0, X1 = X[:, 0], X[:, 1]
xx, yy = make_meshgrid(X0, X1)
for clf, title, ax in zip(models, titles, sub.flatten()):
plot_contours(ax, clf, xx, yy,
cmap=plt.cm.coolwarm, alpha=0.8)
ax.scatter(X0, X1, c=y, cmap=plt.cm.coolwarm, s=20, edgecolors='k')
ax.set_xlim(xx.min(), xx.max())
ax.set_ylim(yy.min(), yy.max())
ax.set_xlabel('Sepal length')
ax.set_ylabel('Sepal width')
ax.set_xticks(())
ax.set_yticks(())
ax.set_title(title)
plt.show()
Automatically created module for IPython interactive environment
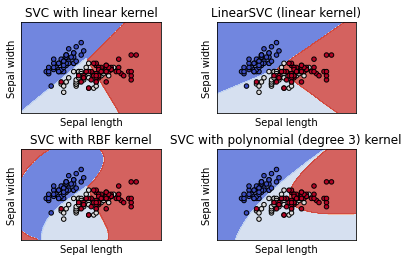
RBF Kernel 함수는 SVM Kernel 함수들 중에 선형 비선형 데이터 모두에서 성능이 좋아 일반적으로 많이 사용됩니다.
주요 파라미터
C: 값이 클수록, Class내에 오류를 허용 하지 않음
Gamma: 값이 클수록, 결정경계 (Decision Boundary) 가 정교해짐
두 파마리터 모두 값이 클수록 학습용 데이터에 대한 분류 결정 경계는 명확해지지만 과적합(Overfitting) 가능성이 커지게 됩니다.
from sklearn import datasets, model_selection, svm, metrics
# 데이터
n_samples = 10000
xs, ys = datasets.make_classification(
n_samples=n_samples, # 데이터 수
n_features=10, # X feature 수
n_informative=3,
n_classes=3, # Y class 수
random_state=rseed) # 난수 발생용 Seed 값
print(f"data shape: xs={xs.shape}, ys={ys.shape}")
train_xs, test_xs, train_ys, test_ys = model_selection.train_test_split(
xs, ys, test_size=0.3, shuffle=True, random_state=2)
print(f"train shape: train_xs={train_xs.shape}, train_ys={train_ys.shape}")
print(f"test shape: test_xs={test_xs.shape}, test_ys={test_ys.shape}")
# 모델
models = [
svm.SVC(kernel='linear', C=1),
svm.SVC(kernel="poly", degree=3, coef0=1, C=5),
svm.SVC(kernel='rbf', decision_function_shape='multinomial')
]
for model in models:
# 학습
print(f"model={model}")
model.fit(train_xs, train_ys)
# 평가
pred_ys = model.predict(test_xs)
acc = metrics.accuracy_score(test_ys, pred_ys)
print(f"acc={acc:.5f}")
cr = metrics.classification_report(test_ys, pred_ys)
print(f"classification_report\n{cr}")
data shape: xs=(10000, 10), ys=(10000,)
train shape: train_xs=(7000, 10), train_ys=(7000,)
test shape: test_xs=(3000, 10), test_ys=(3000,)
model=SVC(C=1, kernel='linear')
acc=0.76700
classification_report
precision recall f1-score support
0 0.80 0.71 0.75 997
1 0.68 0.81 0.74 993
2 0.84 0.78 0.81 1010
accuracy 0.77 3000
macro avg 0.77 0.77 0.77 3000
weighted avg 0.78 0.77 0.77 3000
model=SVC(C=5, coef0=1, kernel='poly')
acc=0.85467
classification_report
precision recall f1-score support
0 0.91 0.84 0.87 997
1 0.77 0.90 0.83 993
2 0.91 0.82 0.86 1010
accuracy 0.85 3000
macro avg 0.86 0.85 0.86 3000
weighted avg 0.86 0.85 0.86 3000
model=SVC(decision_function_shape='multinomial')
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
<ipython-input-3-10ab132992fe> in <module>
25 # 학습
26 print(f"model={model}")
---> 27 model.fit(train_xs, train_ys)
28
29 # 평가
~/.pyenv/versions/3.8.9/envs/skp-n4e-jupyter-sts/lib/python3.8/site-packages/sklearn/svm/_base.py in fit(self, X, y, sample_weight)
159 if hasattr(self, 'decision_function_shape'):
160 if self.decision_function_shape not in ('ovr', 'ovo'):
--> 161 raise ValueError(
162 f"decision_function_shape must be either 'ovr' or 'ovo', "
163 f"got {self.decision_function_shape}."
ValueError: decision_function_shape must be either 'ovr' or 'ovo', got multinomial.


